rstoolbox.plot.multiple_distributions¶
-
rstoolbox.plot.multiple_distributions(df, fig, grid, igrid=None, values='*', titles=None, labels=None, refdata=None, ref_equivalences=None, violins=True, legends=False, **kwargs)¶ Automatically plot boxplot distributions for multiple score types of the decoy population.
A part from the fixed options, the function accepst any option of
boxplot(), except fory,dataandax, which are used internally by this function.Parameters: - df (
DataFrame) – Data container. - fig (
Figure) – Figure into which the data is going to be plotted. - grid (
tuplewith twoint) – Shape of the grid to plot the values in the figure (rows x columns). - igrid (
tuplewith twoint) – Initial position of the grid. Defaults to (0, 0) - values (
list()ofstr) – Contents from the data container that are expected to be plotted. - titles (
list()ofstr) – Titles to assign to the value of each plot (if provided). - labels (
list()ofstr) – Y labels to assign to the value of each plot. By default this will be the name of the value. - refdata (
DataFrame) – Data content to use as reference. - ref_equivalences (dict) – When names between the query data and the provided data are the
same, they will be directly assigned. Here a dictionary
db_name:query_namecan be provided otherwise. - violins (bool) – When
True, plot refdata comparisson with violins, otherwise do it with kdplots. - legends (bool) – When
True, show the legends of each axis.
Returns: list()ofAxesRaises: ValueError: If columns are requested that do not exist in the DataFrame.ValueError: If the given grid does not have enought positions for all the requested values. ValueError: If the number of values and titles do not match. ValueError: If the number of values and labels do not match. ValueError: If refdatais notDataFrame.Example 1: Raw design population data.
In [1]: from rstoolbox.io import parse_rosetta_file ...: from rstoolbox.plot import multiple_distributions ...: import matplotlib.pyplot as plt ...: df = parse_rosetta_file("../rstoolbox/tests/data/input_2seq.minisilent.gz") ...: values = ["score", "hbond_sr_bb", "B_ni_rmsd", "hbond_bb_sc", ...: "cav_vol", "design_score", "packstat", "rmsd_drift"] ...: fig = plt.figure(figsize=(25, 10)) ...: axs = multiple_distributions(df, fig, (2, 4), values=values) ...: plt.tight_layout() ...: In [2]: plt.show() In [3]: plt.close()
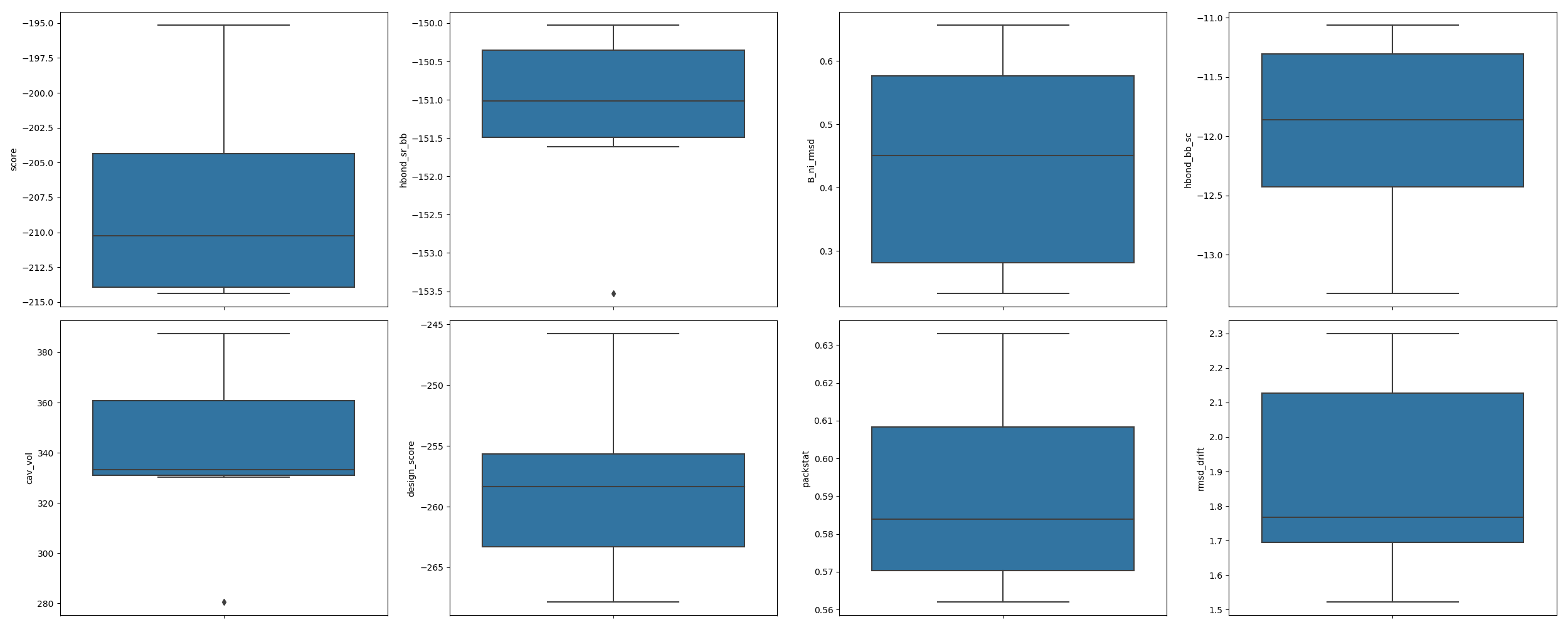
Example 2: Design population data vs. DB reference.
In [4]: from rstoolbox.io import parse_rosetta_file ...: from rstoolbox.plot import multiple_distributions ...: from rstoolbox.utils import load_refdata ...: import matplotlib.pyplot as plt ...: df = parse_rosetta_file("../rstoolbox/tests/data/input_2seq.minisilent.gz", ...: {'sequence': 'A'}) ...: slength = len(df.iloc[0]['sequence_A']) ...: refdf = load_refdata('scop2') ...: refdf = refdf[(refdf['length'] >= slength - 5) & ...: (refdf['length'] <= slength + 5)] ...: values = ["score", "hbond_sr_bb", "B_ni_rmsd", "hbond_bb_sc", ...: "cav_vol", "design_score", "packstat", "rmsd_drift"] ...: fig = plt.figure(figsize=(25, 10)) ...: axs = multiple_distributions(df, fig, (2, 4), values=values, refdata=refdf) ...: plt.tight_layout() ...: In [5]: plt.show() In [6]: plt.close()
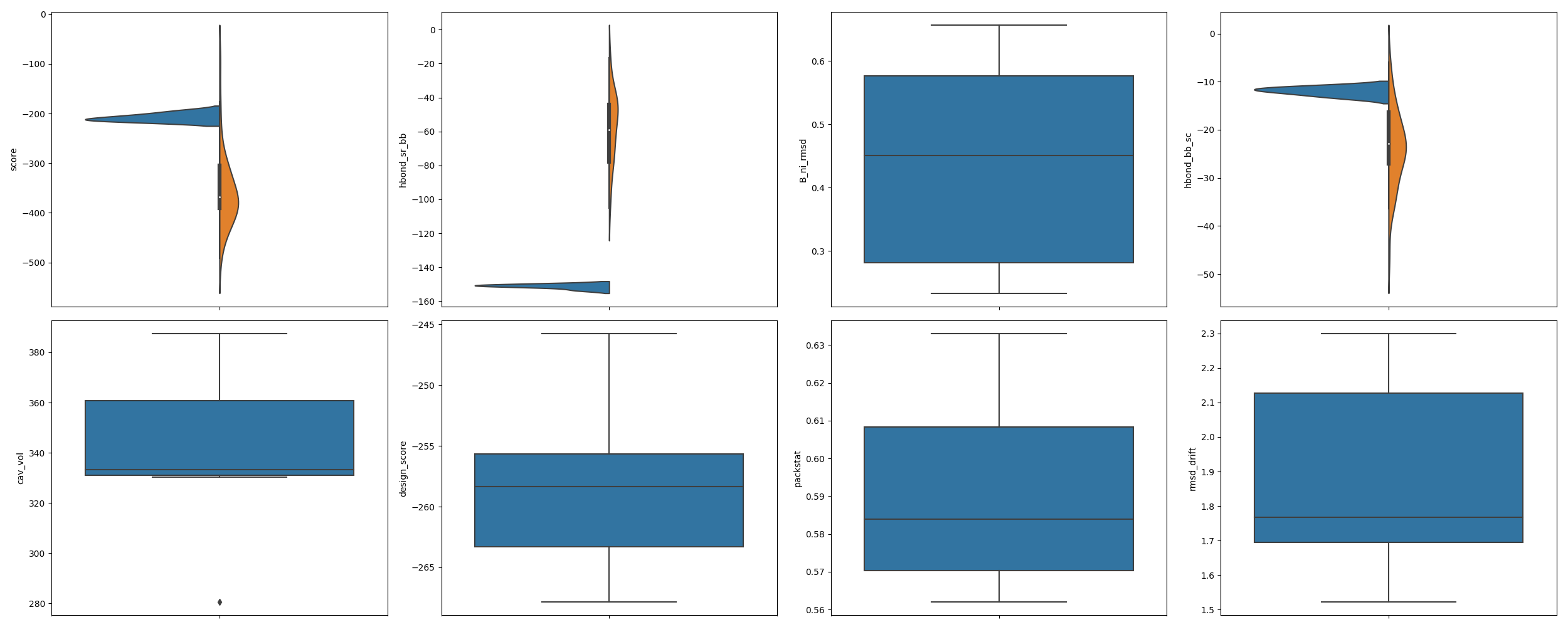
- df (